Homology modeling In
discovery Studio
üExperimental
structure determination involves difficult methods that require a significant
amount of expertise and resources, and can take years to yield results.
ü Protein Modeling
on the other hand enables access to sensible structural models within a matter
of hours or even minutes.
üTemplate based
modeling has been shown to yield useful models in countless publications. As
more and more experimental structures become available, the greater the scope
template based Protein Modeling will provide researchers with accurate models
quickly and efficiently.
üProtein Modeling
and Sequence Analysis solutions in Discovery Studio provide the necessary set
of tools for the construction of molecular structures, as well as
macromolecular docking, in an easy to use customizable graphical user interface
suitable for novice and expert users.
Pre Requirement for Homology Modeling.
ØFirst of all find
the protein sequence which you want to make 3D.
ØThis sequence is
known as a model sequence.
ØKnow download that
template sequence in .PDB format.From Here
ØNow convert model
sequence in .PIR format by copy the sequance in on notepad
file and save as a .pir format.
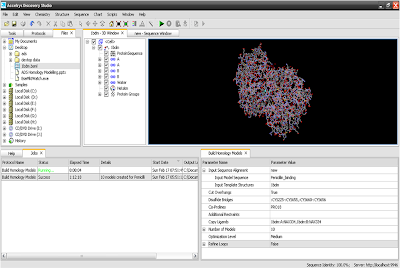
ØNow go through
below steps here pennicilin binding is our model protein sequance and 1btm.PDB is our known protein.
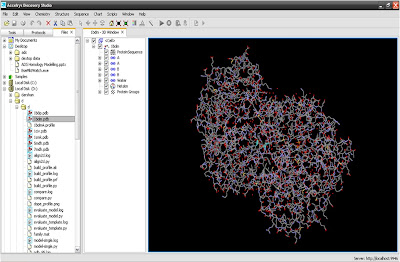
1. Insert template protein structure
2. Find out the sequence from protein structure
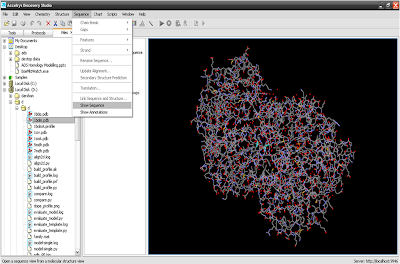
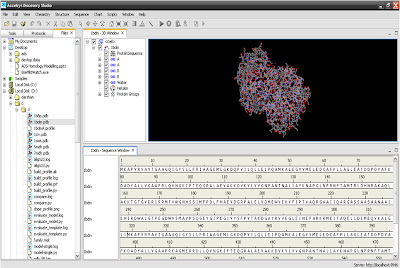
3. Sequence of protein structure
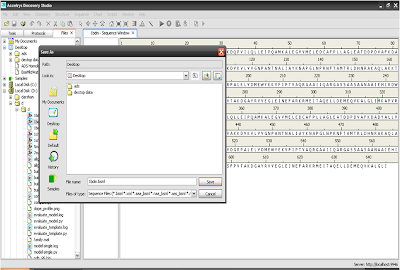
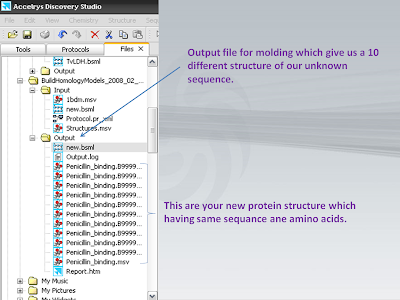
4. Save that sequence as a .bsml formate
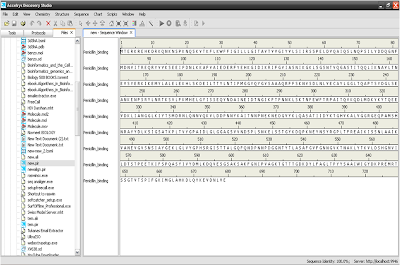
5. Open model sequence in new windows.
6. Drag that template sequence on the model sequence.
7. Open template structure.
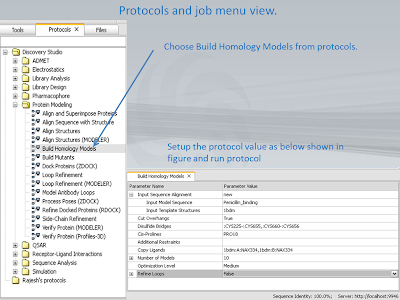
8. Protocols and job menu view.
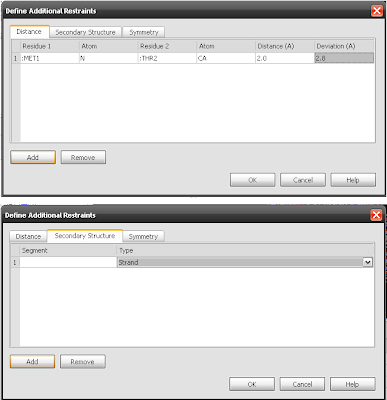
9. Menu for additional restrians
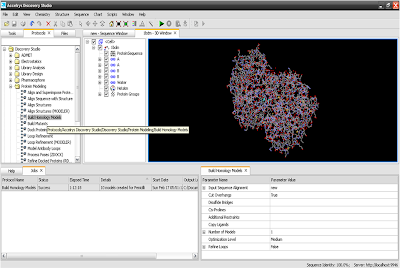
10. Click on run to run protocol.
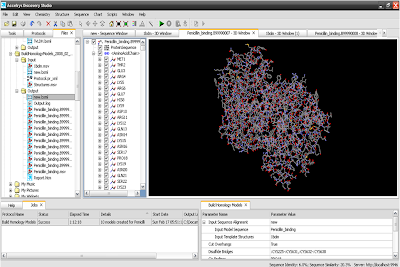
11. Here is your output.

No comments:
Post a Comment